Functional Mechanism
Assemble
Mechanism
Our NanoMuscles are composed of two monomers, “BIG” monomer and “SMALL” monomer. Only when two monomers combine correctly can this DNA origami muscle functions completely. The following section describes how Big monomer and Small monomer assemble as we desire.
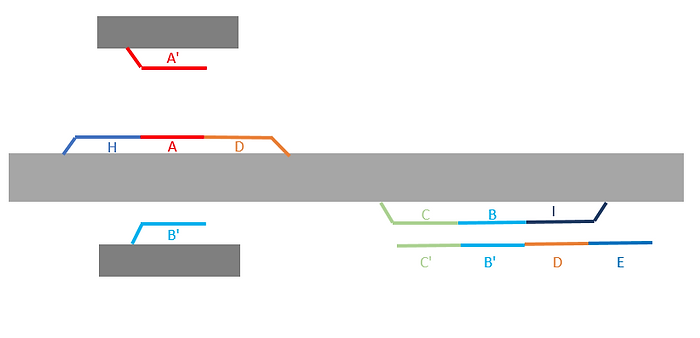
Initially, we designed a lot of single-strand DNA sequences protruding from the Big monomer and Small monomer. After folding process, the structure of open-ring Big monomer and open-ring Small monomer with protruding staples were expected as shown in Figure F1.

Figure F1. Big monomer and Small monomer. Colorful fragments stand for staples that protrude from the main structure (gray).
Before assembly of Big monomer and Small monomer, “blocking(protecting) staples” was added to Big monomer to avoid staple HAD and staple CBI interacting with staple A’ and staple C’ on Small monomer shown as Figure F2.
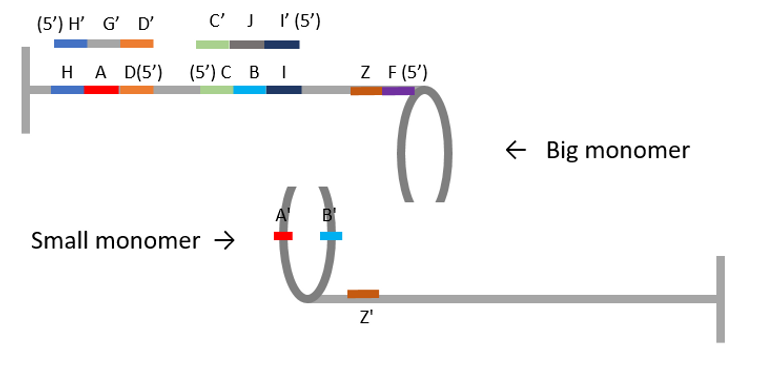
Figure F2. Add blocking staples (H’G’D’ and C’JI’).
After we added blocking staples, Big monomer and Small monomer were mixed to assemble NanoMuscles.
Staple Z and staple Z’ were dimer combining sequences that guided Big monomer and Small monomer to bind to each other in correct orientation. After annealing (about 2~3 days), the ring-closing staples was added in order to close the ring. Then, the interlocked structure was done. (Figure F3)


Figure F3. Dimerization of Big and Small monomers.
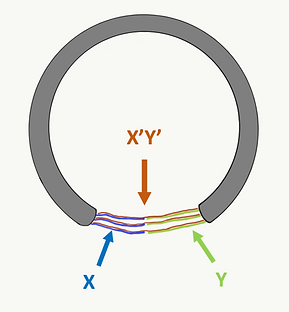
At each end of open-ring, we designed several single-stranded DNA staples for ring-closing (X and Y). The ring-closing staples (X’Y’) are perfectly complementary to the single-stranded DNA of both ends. (Figure F4a and Figure F4b)

Figure F4a. The half-circle represents the open-ring of our design.
Figure F4b. Closed ring after we add ring-closing staples.
Before the function of NanoMucsle was executed, staple HGD and staple CJ’I was added in order to remove the blocking staples and de-block the two main staples (HAD and CBI) on the Big monomer. On the other hand, staple Z’F’ was also added to block the dimer combining sequence to free two monomers (still interlocked). (Figure F5)
Figure F5. Anti-blocking and Free the two monomers (still interlocked).
In this stage, the assembly of NanoMuscle wass complete. The next section will introduce how NanoMuscle displays its function.
In order to make NanoMucsle contract or extend, we add the "Fuel" or "Anti-fuel" into the reaction. In our design, shown in Figure F6, there are sequences A’ and B’ on the ring, and sequences HAD (contraction sequence) and CBI (extension sequence) on the axle. In the contracted state, the protective staple C’B’DE (also called Fuel) anneals to the extension sequence, preventing staple B’ from binding to extension sequence.
Figure F6. Ring structure on staple HAD (contraction sequence). Each letter stands for unique staple sequence, and follows the rule that letter and prime-letter are complementary to each other (Ex, A and A’).
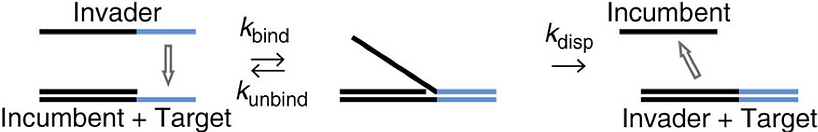
When “Anti-fuel”, of which sequences are CA’D’E’, is added for a while, staple A’ is separated from contraction sequence (Figure F7) due to toehold-mediated strand displacement (Figure F8). The principle is that the more complementary base pairs exist in two single-stranded DNA sequences, the stronger binding tendency it is.
Figure F7. Adding Anti-fuel.
According to Figure F7, staple A’ and contraction sequence (HAD) have only one complementary section, while staple CA’D’E’ and contraction sequence (HAD) have two complementary sections. So, “Anti-fuel” replaces the original staple A’. Similarly, staple C’B’DE and staple extension sequence (CBI) have two complementary sections, while staple CA’D’E’ and staple C’B’DE have three complementary sections. Therefore, “Anti-fuel” replaces the protecting staple C’B’DE.
In the next step (Figure F9), NanoMuscle start extending. The ring shifts to the position where staple B’ and staple extension sequence (CBI) bind to each other.
Figure F8. Toehold-mediated strand displacement. [F-1]
Figure F9. Ring structure on extension sequence (CBI).
Fuel is the counter partner of Anti-fuel. It functions similar mechanism as Anti-fuel. After Fuel is added in reaction, Anti-fuel detaches from monomers of NanoMucsle, and the staple A on the ring is capable of interacting with the staple A’. At the same time, extension sequence (CBI) is blocked by fuel and the ring is freed. (Figure F10)
Figure F10. Adding Fuel.
In the end, NanoMucscle changes back to the contracted state due to the interaction between staple contraction sequence(HAD) and A’. (Figure F6)
To sum up, NanoMucsle is a controllable nanomachine that can extend or contract by supplying Anti-fuel or Fuel.
Citation:
[F-1]
Machinek, Robert RF, et al. "Programmable energy landscapes for kinetic control of DNA strand displacement." Nature communications 5 (2014): 5324.